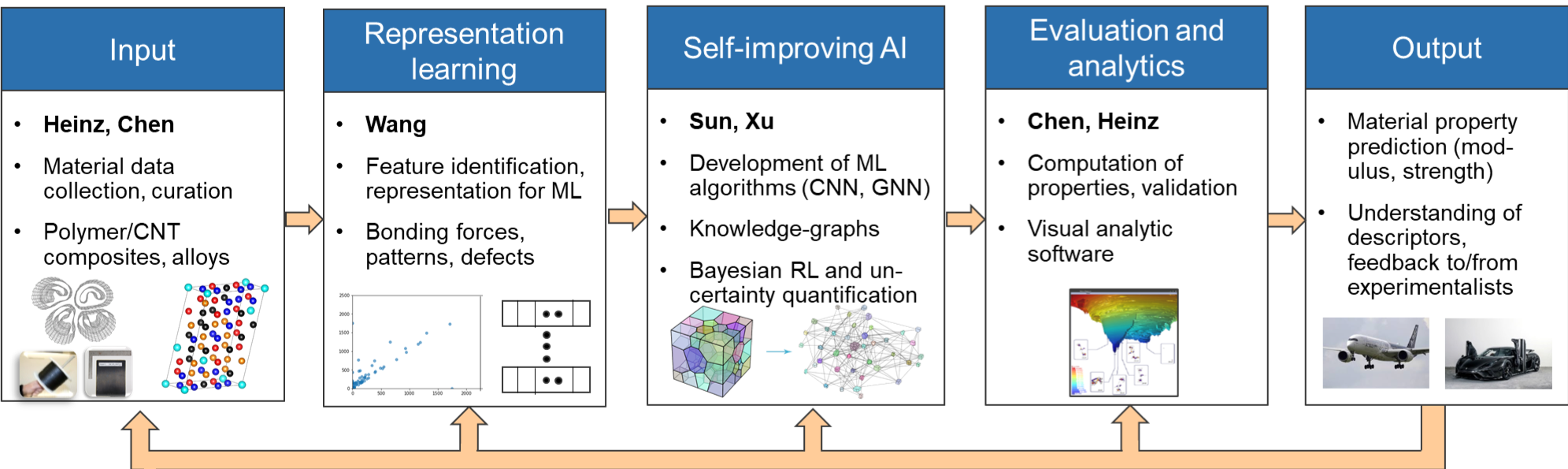
Methods and applications to HIV related studies
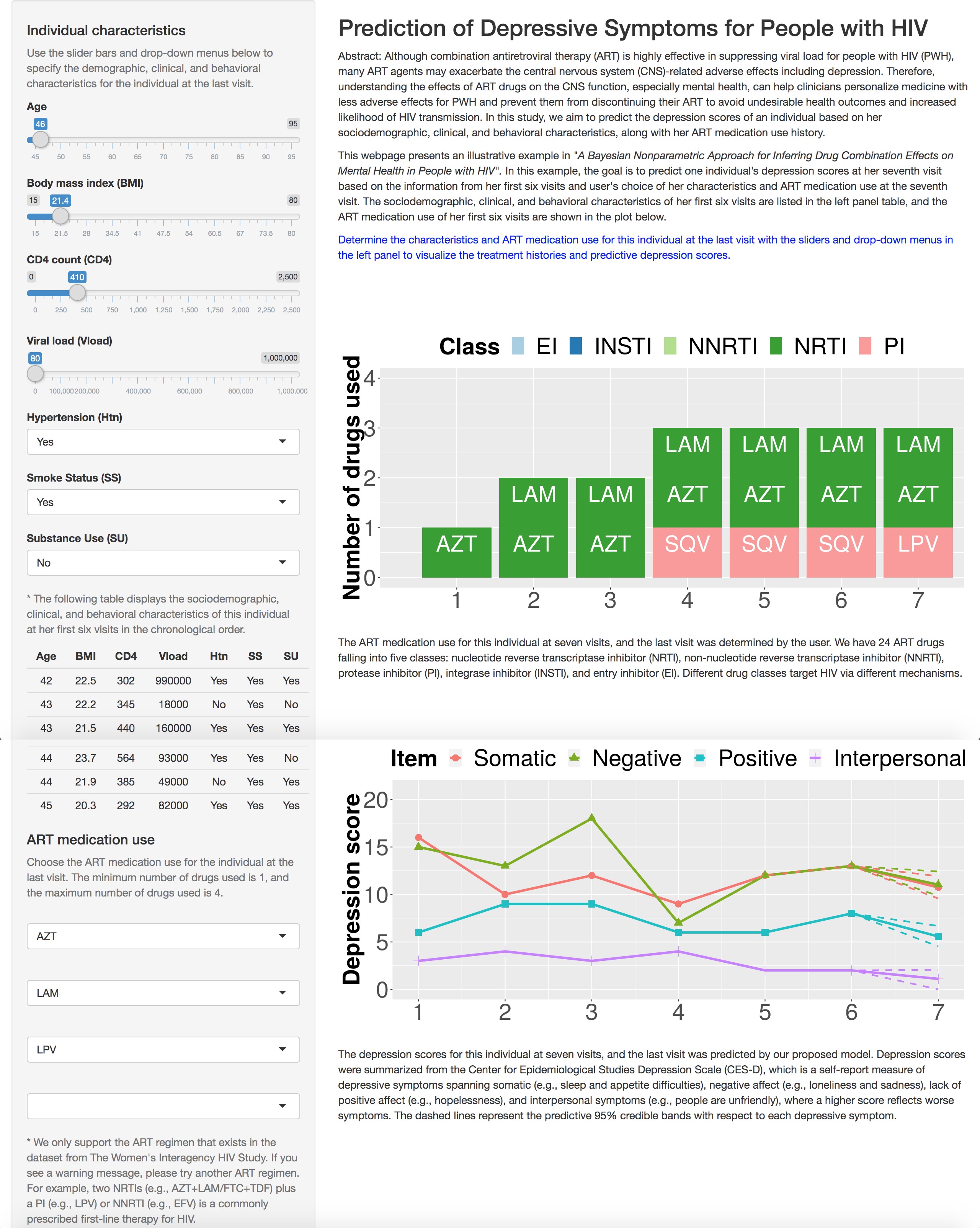
Although combination antiretroviral therapy (ART) is highly effective in suppressing viral load for people with HIV (PWH), many ART agents may exacerbate central nervous system (CNS)-related adverse effects including depression. Therefore, understanding the effects of ART drugs on the CNS function, especially mental health, can help clinicians personalize medicine with less adverse effects for PWH and prevent them from discontinuing their ART to avoid undesirable health outcomes and increased likelihood of HIV transmission. The emergence of electronic health records offers researchers unprecedented access to HIV data including individuals’ mental health records, drug prescriptions, and clinical information over time. However, modeling such data is very challenging due to high-dimensionality of the drug combination space, the individual heterogeneity, and sparseness of the observed drug combinations. We develop Bayesian approaches to learn longitudinal drug effects and drug combination effects on mental health in PWH adjusting for socio-demographic, behavioral, and clinical factors. Our method has clinical utility in guiding clinicians to prescribe more informed and effective personalized treatment based on individuals’ treatment histories and clinical characteristics.
Selected Publications:
- Jin W+ , Ni Y, Rubin LH, Spence AB and Xu Y# , “A Bayesian Nonparametric Approach for Inferring Drug Combination Effects on Mental Health in People with HIV.” arXiv:2004.05487 / RShiny
- Asante R Kamkwalala* , Kunbo Wang*+ , P Jane O’Halloran, Dionna W. Williams, Raha Dastgheyb, Kathryn C. Fitzgerald, Amanda B. Spence, Pauline M. Maki, Deborah R. Gustafson, Joel Milam, Anjali Sharma, Kathleen M. Weber, Adaora A. Adimora, Igho Ofotokun, Anandi N. Sheth, Cecile D. Lahiri, Margaret A. Fischl, Deborah Konkle-Parker, Xu Y# , Rubin LH# , “Higher peripheral monocyte activation markers are
associated with smaller frontal and temporal cortical volumes in women with HIV”.
AIDS and Behavior, 2020.
Selected Grants:
- NSF 1918854 (Principal Investigator)
- The Johns Hopkins Center for AIDS Research Faculty Development Award (Principal Investigator)
- NIH R01MH120693 (Co-Investigator)
- NIH R01MH119947 (Co-Investigator)
- NIH R01MH113512 (Co-Investigator)
Selected Collaborators:
- Leah H. Rubin, Ph.D. , Johns Hopkins School of Medicine.
- Yang Ni, Ph.D. , Texas A&M University.
- Dionna W Williams, Ph.D. , Johns Hopkins School of Medicine.
- Jane O’Halloran, MD , Washington University School of Medicine in St.Louis.